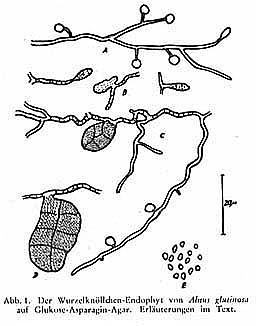
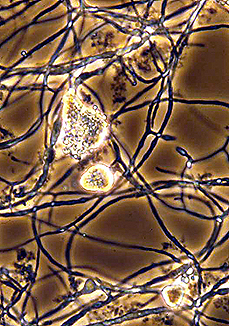
The first Frankia isolate was reported in 1959 by Ernst-Heinrich Pommer working at the Botanischen Institut der Technischen Hochschule Aachen. (Left). CpI1 spores and hyphae (Right, phase contrast; D. Benson).
Actinomycete taxonomy has traditionally been based on morphology, "chemotaxonomy" and, more recently, on phylogenetic analysis. The morphology of Frankia is discussed here. The genus Frankia was named originally by Brunchorst in 1886 to honor A. B. Frank, a Swiss microbiologist. Brunchorst at the time considered the organism living in the root nodules to be a filamentous fungus. Becking (1970) redefined the genus as containing prokaryotic actinomycetes and created the family Frankiaceae within the Actinomycetales (now Actinobacteria). No isolates were available at that time, although Pommer had most likely obtained one (above), and the organisms were considered to be "obligate" symbionts. Nevertheless, Becking's acceptance of the genus Frankia has remained and continues to be used.
At present, the genus Frankia is classified as follows in Bergey's Manual:
| Phylum BXIV | Actinobacteria | |
| Class I | Actinobacteria | |
| Subclass V | Actinobacteridae | |
| Suborder XIII | Frankineae | |
| Family I | Frankiaceae | |
| Genus | Frankia |
Thus, it is fair to say that the organism has much in common with other actinobacteria, including members of the Corynebacteriaceae, Streptomycetaceae and Mycobacteriaceae. Analysis of 16S ribosomal RNA genes reveals that frankiae form a coherent clade within the Actinobacteria. Information on phylogenetic analyses can be found here.
Chemotaxonomy
Chemotaxonomy refers to grouping organisms according to similarities of cell chemistry including wall constituents, membranes and quinones. G+C ratios are also important but cannot be used for species identification because all Frankia as well as other actinobacteria have high Mol% G+C.
The G + C content of Frankia strains is around 66-75 Mol % which is typical of the filamentous Actinobacteria (An, 1983; Fernandez, 1989). Frankia strains have a Type III cell wall (meso-diaminopimelic acid, alanine, glutamic acid, muramic acid and glucosamine (Lechevalier, 1990)) commonly found amongst aerobic actinomycetes. Whole cell sugars include fucose, ribose, xylose, madurose, mannose, galactose, glucose, rhamnose, and 2-O-methyl-D-mannose (Lechevalier, 1990; Mort, 1983; St-Laurent, 1987). Phospholipid analysis places Frankia strains into group PI containing phosphatidyl inositol, phosphatidyl inositol mannosides, and diphosphatidyl glycerol; nitrogen-containing phospholipids are absent (Lechevalier, 1989). This grouping distinguishes them from Geodermatophilus (group PII) which also produce multilocular sporangia, and were once considered close relatives of Frankia (Hahn, 1989). In the few strains that have been analyzed, a menaquinone pattern consisting mainly of MK9 (H4) (menaquinone with nine isoprene units, four of which are hydrogenated) with lesser amounts of MK9 (H6 and H8) was found (Lechevalier, 1987).
Frankia species?
As minimum criteria for including strains within the genus, morphology in culture, together with isolation from an actinorhizal root nodule, and usually a analysis of the 16S ribosomal RNA gene are sufficient (Lechevalier, 1989, 1990). Designating species has been problematical (Lechevalier, 1989). Becking (1970) named species according to plants infected. This approach suffers from the fact that some strains can nodulate plants from different plant orders and some strains cannot reinfect their source plant (Baker, 1987; Bosco, 1992; An, 1985; Fernandez, 1989). In addition, broad patterns of host plant specificity have been observed in ecological studies of host and symbiont distribution (see Frankia Ecology).
The advent of several Frankia genome sequences has revealed wide fluctuations in sizes, despite a divergence in the 16S rRNA genes that, by traditional standards, might place all strains in the same species (>97% sequence identity). Nevertheless several species have been named based on plant source or infectivity (Gtari et al. 2019). These include Frankia alni, F. torreyi, F. canadensis, F. casuarinae, F. coriariae, F. elaeagni, F. discariae, F. irregularis, F. inefficax, F. asymbiotica,, and F. saprophytica,